···
Mungo Carstairs
Jalview Computational Scientist
The Barton Group
Division of Computational Biology
School of Life Sciences
University of Dundee, Dundee, Scotland, UK.
www.jalview.org
www.compbio.dundee.ac.uk
From: Mungo Carstairs (Staff)
Sent: 16 November 2016 13:45:52
To: Geoffrey Barton (Staff)
Subject: Re: [Jalview-dev] More Jalview-Chimera goodness
spike-mungo updated - now ignores ‘False’ attribute values in Chimera.
Which means isHelix feature is only added where, well, the residue is helix.
For now, features are being added at a single locus only.
A refinement may be to set as a range e.g. (FER2_ARATH) isHelix:75-83 etc.
mungo
The University of Dundee is a registered Scottish Charity, No: SC015096
Mungo Carstairs
Jalview Computational Scientist
The Barton Group
Division of Computational Biology
School of Life Sciences
University of Dundee, Dundee, Scotland, UK.
www.jalview.org
www.compbio.dundee.ac.uk
From: Mungo Carstairs (Staff)
Sent: 15 November 2016 16:56:19
To: Geoffrey Barton (Staff)
Subject: Re: [Jalview-dev] More Jalview-Chimera goodness
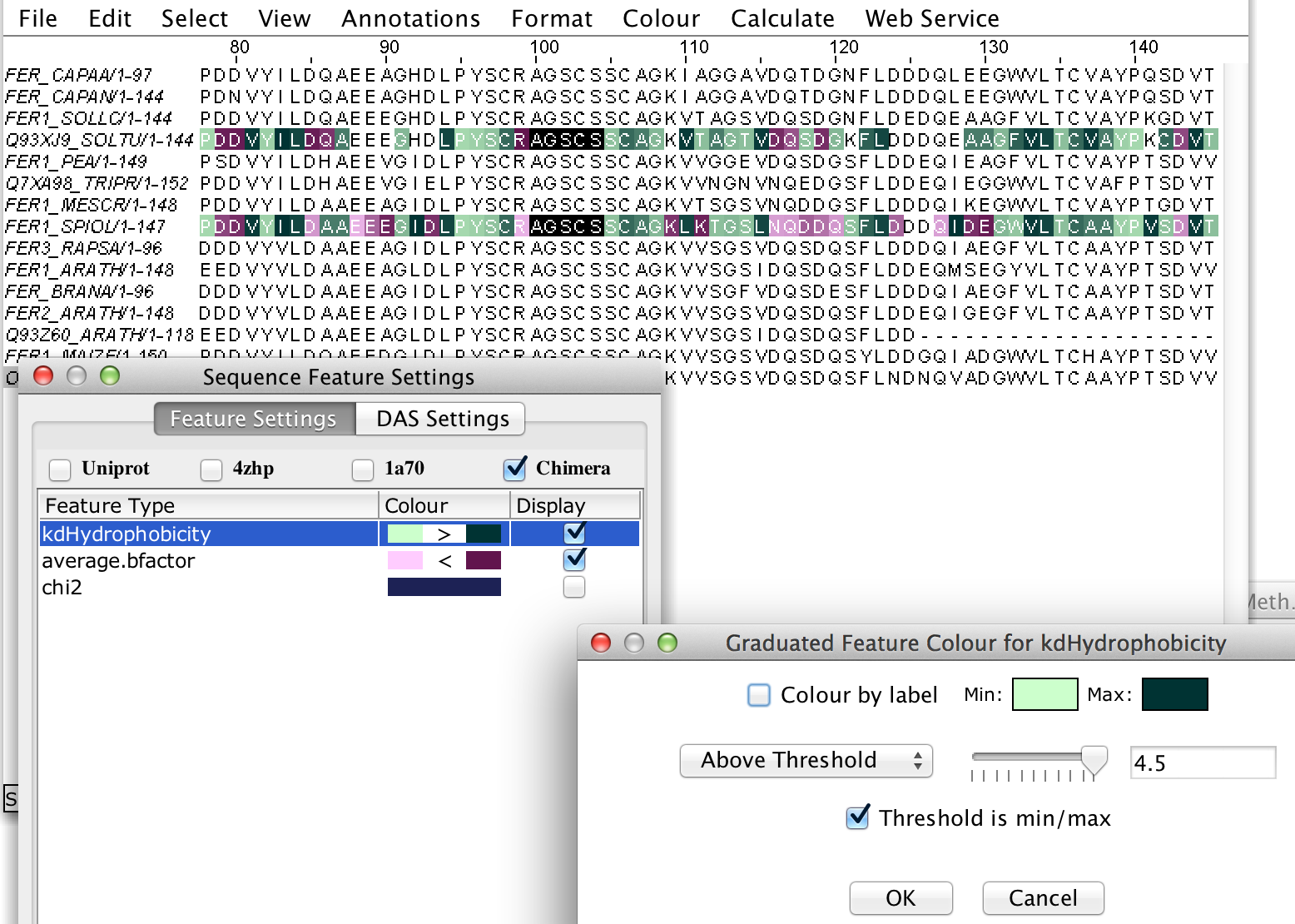
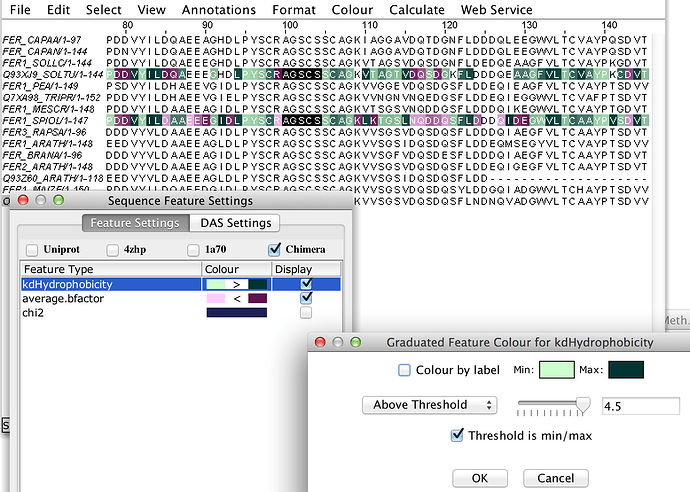
To clarify my answer…Jalview doesn’t automatically make a graduated colour, you have to configure this yourself in Feature Setttings (right click on feature colour, choose Graduated Colour, etc).
It might be possible to default to a graduated colour scheme if a numeric valued feature is detected.
m
Mungo Carstairs
Jalview Computational Scientist
The Barton Group
Division of Computational Biology
School of Life Sciences
University of Dundee, Dundee, Scotland, UK.
www.jalview.org
www.compbio.dundee.ac.uk
From: Mungo Carstairs (Staff)
Sent: 15 November 2016 16:54:41
To: Geoffrey Barton (Staff)
Subject: Re: [Jalview-dev] More Jalview-Chimera goodness
I think I see…
isHelix has values True or False in Chimera (rather than attribute present or absent).
This simplistically transfers to a Jalview feature ‘isHelix’ and every position has it!
I should probably filter out the False values when setting features.
I can set a graduated, thresholded colour for average.bfactor in Jalview though.
mungo
p.s. leaving shortly but can pick up in the morning
Mungo Carstairs
Jalview Computational Scientist
The Barton Group
Division of Computational Biology
School of Life Sciences
University of Dundee, Dundee, Scotland, UK.
www.jalview.org
www.compbio.dundee.ac.uk
From: Geoffrey Barton (Staff)
Sent: 15 November 2016 16:23:02
To: Mungo Carstairs (Staff)
Subject: Re: [Jalview-dev] More Jalview-Chimera goodness
Ah Ok. thanks for the pointer.
Although the chimera attributes are getting transferred, but colouring is fixed at the whole domain rather than the region. I tried average.bfactor first and expected this to give me a colour ramp since I assumed average.bfactor was the bfactor averaged for the atoms in each residue rather than over the whole molecule. I then tried is.Helix and is.Strand and these just changed the colour of the whole molecule.
Geoff.
On 15/11/2016 15:50, Mungo Carstairs (Staff) wrote:
Hi Geoff,
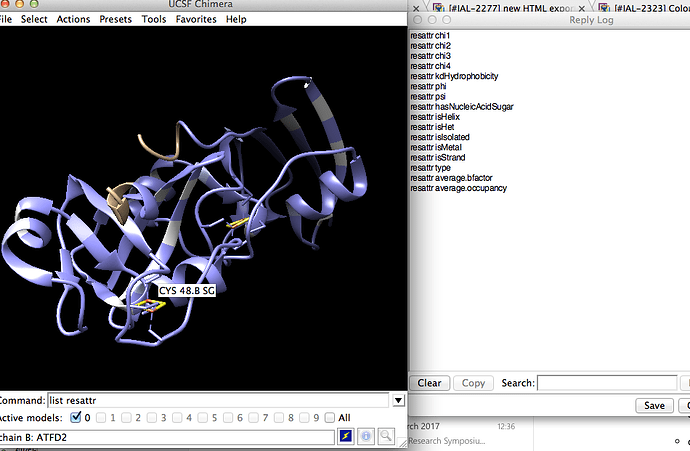
In Chimera, menu option Favorites | Reply Log opens the reply log, and Favorites | Command Line opens the command line.
![]()
mungo
--
Geoff Barton | Professor of Bioinformatics | Head of Division of Computational Biology
School of Life Sciences | University of Dundee, Scotland, UK | [g.j.barton@dundee.ac.uk](mailto:g.j.barton@dundee.ac.uk)
Tel: +44 1382 385860 | [www.compbio.dundee.ac.uk](http://www.compbio.dundee.ac.uk) | twitter: @gjbarton
The University of Dundee is registered Scottish charity: No.SC015096
Mungo Carstairs
Jalview Computational Scientist
The Barton Group
Division of Computational Biology
School of Life Sciences
University of Dundee, Dundee, Scotland, UK.
www.jalview.org
www.compbio.dundee.ac.uk
From: Geoffrey Barton (Staff)
Sent: 15 November 2016 15:44:53
To: Mungo Carstairs (Staff)
Subject: Re: [Jalview-dev] More Jalview-Chimera goodness
HI Mungo,
Just looking at this. Got stuck…
On 15/11/2016 09:24, Mungo Carstairs (Staff) wrote:
http://www.jalview.org/builds/spike-mungo/webstart/jalview_2G.jnlp
has a prototype of:
To demonstrate, set up with
-
Input alignment from URL http://www.jalview.org/examples/uniref50.fa
-
Web Services | Fetch DB Refs | from Uniprot to add features
-
View | Show Sequence Features
-
View a structure in Chimera for FER1_SPIOL
Now you can:
-
from the Chimera View panel, select “Chimera | Write Jalview features”
- this copies any visible features to Chimera attributes (except for RESNUM) on mapped residue positions
- to see the new attributes, open the Chimera reply log and command line, and type “list resattr” and “list resi attr jv_domain”
Where is the Chimera reply log?
-
- attribute names are given a “jv_” prefix as a ‘Jalview namespace’
- now View | Feature Settings in Jalview, and from the Chimera View panel select “Chimera | Fetch Chimera attributes”
- a sub-menu with a list of Chimera residue attributes should be offered
- (this occasionally pauses or hangs as it queries Chimera…if so try again…)
- click on an attribute name to copy it to Jalview - it should magically appear in the Feature Settings, under group “Chimera”
- you can now colour / select on the feature in the usual (Jalview) way
- similarly, numeric valued attributes (e.g. phi, chi1, kdHydrophobicity etc) can have a graduated and/or thresholded colour applied
![]()
Note: not merged on to this version (but could be): option added by Jim for ‘create selection from highlight’ in Jalview after selecting in Chimera.
Any feedback on this welcome.
The main remaining related enhancement for this (so far!) is JAL-2294 - support for combining annotations in Jalview into ‘derived’ annotation.
That may take a bit longer…
thanks,
mungo
The University of Dundee is a registered Scottish Charity, No: SC015096
_______________________________________________
Jalview-dev mailing list
[Jalview-dev@jalview.org](mailto:Jalview-dev@jalview.org)
[http://www.compbio.dundee.ac.uk/mailman/listinfo/jalview-dev](http://www.compbio.dundee.ac.uk/mailman/listinfo/jalview-dev)
--
Geoff Barton | Professor of Bioinformatics | Head of Division of Computational Biology
School of Life Sciences | University of Dundee, Scotland, UK | [g.j.barton@dundee.ac.uk](mailto:g.j.barton@dundee.ac.uk)
Tel: +44 1382 385860 | [www.compbio.dundee.ac.uk](http://www.compbio.dundee.ac.uk) | twitter: @gjbarton
The University of Dundee is registered Scottish charity: No.SC015096
Mungo Carstairs
Jalview Computational Scientist
The Barton Group
Division of Computational Biology
School of Life Sciences
University of Dundee, Dundee, Scotland, UK.
www.jalview.org
www.compbio.dundee.ac.uk