Previously (about 4 months ago) I was able to colour the sequence alignment according to conservation + a selected colour scheme (e.g. zappo), or show colours according to identity threshold, and if I view the protein structure (using local file) in PyMOL the colouring of the sequence alignment could be applied in the structure. Now I can’t seem to do this anymore - the structure can be coloured according to any colour scheme, but conservation or identity threshold information are not applied in PyMOL.
Hi there.
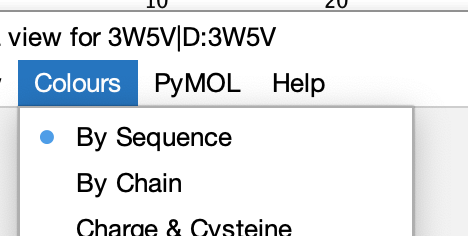
Could you make sure that ‘By Sequence’ is selected in the Jalview desktop’s Pymol window’s Colours menu ?
If that’s selected, then any shading applied to the linked sequence in the alignment should also be faithfully applied to the Pymol view.
You can also check that everything is working correctly via the Jalview Java Console (opened via the Tools menu). If there are any errors or exceptions appearing in the console when you open the Pymol view, these may point to a problem. You can also adjust the log level to ‘Trace’ to show all the commands as they are sent to pymol.
If everything seems to check out, then send me your files so I can take a look at whats going on - either direct to my dundee.ac.uk email (jprocter) or as a DM on discourse.
hear from you soon !
Jim
Clicking “By Sequence” repeatedly resulting in nothing. Other colour options seem to function as expected
When by sequence is ticked, you should then be able to adjust the colours in the aligment view(s) linked to the pymol view. Repeated selection will not necessarily update the structure view(s) colours more than once.
Regardless, if once you have by sequence ticked and have applied a colour scheme and en/disabled conservation or PID shading on the alignment (and that shading actually changed the appearance of the sequence linked to the structure in the view), then the structure view should update. If it doesn’t,
please send me an example to test, and also the contents of the jalview java console log.
I also need the pymol version you are using, though it sounds like everything there is working fine.
Jim.
The “By Sequence” button is simply taking no action. Whether opening the structure in PyMOL before or after, selecting a colour scheme for the sequence alignment results in no update to the structure. Selecting the colour scheme in the Jalview Pymol Structure Viewer window does update the structure. Jmol has no problem.
PyMOL version 2.5.2. I’m quite sure that this is the same version that I used approx. 4 months ago when I was able to do the described task.
Jalview Console (with several hyperlinks removed)
Jalview Version: 2.11.2.5
Jalview Installation: Source git-commit:519295fc04 [releases/Release_2_11_2_Branch]
Build Date: 16:54:56 11 October 2022
Java version: 1.8.0_322
amd64 Windows 10 10.0
Install4j version: 9.0.5
Install4j template version: 2.11.2.0_F188884b4_C2040e7e728
Launcher version: 1.8.3-1.2.12_JVL
LookAndFeel: Windows (com.sun.java.swing.plaf.windows.WindowsLookAndFeel)
INFO - Cannot set Taskbar
ERROR - java/awt/Taskbar
File format identified as Fasta
https://www.ebi.ac.uk/pdbe/search/pdb/select?wt=json&fl=pdb_id,title,experimental_method,resolution&rows=500&start=0&q=(text:101)+AND+molecule_sequence:[‘’+TO+*%5D+AND+status:REL&sort=overall_quality+desc
query >>>>>>> jalview.fts.core.FTSRestRequest@17eaca37
Operating headless display=null nographicsallowed=true
(C) 2015 Jmol Development
Jmol Version: 15.1.53 2021-02-10 09:12
java.vendor: Java: Temurin
java.version: Java 1.8.0_322
os.name: Windows 10
Access: ALL
memory: 109.5/322.4
processors available: 8
useCommandThread: false
The Resolver thinks Pdb
reading 998 atoms
ModelSet: haveSymmetry:false haveUnitcells:false haveFractionalCoord:false
1 model in this collection. Use getProperty “modelInfo” or getProperty “auxiliaryInfo” to inspect them.
Default Van der Waals type for model set to Jmol
998 atoms created
ModelSet: autobonding; use autobond=false to not generate bonds automatically
Jmol 15.1.53 2021-02-10 09:12 DSSP analysis for model 1.1 - null
W. Kabsch and C. Sander, Biopolymers, vol 22, 1983, pp 2577-2637
We thank Wolfgang Kabsch and Chris Sander for writing the DSSP software,
and we thank the CMBI for maintaining it to the extent that it was easy to
re-engineer in Java for our purposes.
Second generation DSSP 2.0 is used in this analysis. See Int. J. Mol. Sci. 2014, 15, 7841-7864; doi:10.3390/ijms15057841.
All bioshapes have been deleted and must be regenerated.
Time for creating model: 16 ms
INFO - PyMOL started with XMLRPC on port 9123
INFO - PyMOL XMLRPC started on port 9123
CIR resolver set to https://cactus.nci.nih.gov/chemical/structure
Operating headless display=null nographicsallowed=true
(C) 2015 Jmol Development
Jmol Version: 15.1.53 2021-02-10 09:12
java.vendor: Java: Temurin
java.version: Java 1.8.0_322
os.name: Windows 10
Access: ALL
memory: 41.9/325.6
processors available: 8
useCommandThread: false
The Resolver thinks Pdb
reading 998 atoms
ModelSet: haveSymmetry:false haveUnitcells:false haveFractionalCoord:false
1 model in this collection. Use getProperty “modelInfo” or getProperty “auxiliaryInfo” to inspect them.
Default Van der Waals type for model set to Jmol
998 atoms created
ModelSet: autobonding; use autobond=false to not generate bonds automatically
Jmol 15.1.53 2021-02-10 09:12 DSSP analysis for model 1.1 - null
W. Kabsch and C. Sander, Biopolymers, vol 22, 1983, pp 2577-2637
We thank Wolfgang Kabsch and Chris Sander for writing the DSSP software,
and we thank the CMBI for maintaining it to the extent that it was easy to
re-engineer in Java for our purposes.
Second generation DSSP 2.0 is used in this analysis. See Int. J. Mol. Sci. 2014, 15, 7841-7864; doi:10.3390/ijms15057841.
All bioshapes have been deleted and must be regenerated.
Time for creating model: 10 ms
PyMOL Console
PyMOL™ 2.5.2 - Incentive Product
Copyright (C) Schrodinger, LLC
This Executable Build integrates and extends Open-Source PyMOL.
Detected OpenGL version 4.6. Shaders available.
Detected GLSL version 4.60.
OpenGL graphics engine:
GL_VENDOR: Intel
GL_RENDERER: Intel(R) UHD Graphics
GL_VERSION: 4.6.0 - Build 30.0.101.1404
License Expiry date: 15-jun-2023
Detected 8 CPU cores. Enabled multithreaded rendering.
xml-rpc server running on host localhost, port 9123
Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b
Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b
Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndi
vidual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b
Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndi
vidual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b
Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndi
vidual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CC
oLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b Selector-Error: Invalid selection name "C_3A_5CUsers_5CTW43969_5COneDrive++University+of+Dundee_5CCurrent_5CResearch_5CTom+W+Research+Record_5CBioinformatics_5CStructure_5CAlphaFold_5CCoLabFold_5CColabFold+Results_5CIndividual+Protein+Results_5CMp1_77b
Thanks for posting the console log - I can see straight away there are errors when importing the structure from your onedrive mounted home directory. This is unfortunately a long standing problem that we have not yet resolved with Jalview, https://issues.jalview.org/browse/JAL-2275
There is a workaround. If you first copy the PDB file to your laptop’s local drive, then load it on to the sequence alignment, it will work fine.