Hello Maitreyi,
Glad to hear you are interested in integrating the Jalview applet to your site!
I ran your example using Safari with ‘Show Java Console’ enabled to see any error messages, and saw:
java.lang.NoClassDefFoundError: org/jmol/api/JmolSelectionListener
Please can you ensure that your applet classpath includes the jar file for Jmol (3d structure viewer).
There is a link to this (as ‘JmolApplet.jar’) on http://www.jalview.org/examples/.
The full jar file name is
JmolApplet-14.2.14_2015.06.11.jar
Download this file and add it to the archive parameter of your tag.
This ought to solve your problem!
Best regards,
Mungo
The University of Dundee is a registered Scottish Charity, No: SC015096
···
Mungo Carstairs
Jalview Computational Scientist
The Barton Group
Division of Computational Biology
School of Life Sciences
University of Dundee, Dundee, Scotland, UK.
www.jalview.org
www.compbio.dundee.ac.uk
From: Geoffrey Barton (Staff)
Sent: 07 July 2016 09:08:58
To: Ashok, Maitreyi (Maitreyi)
Subject: Re: JalviewLite question
Dear Maitreyi,
Sorry to hear you are having troubles with the applet. I’ll forward your message to the team and someone will get back to you.
If you have further problems with Jalview it is best if you post requests for help to the jalview-discuss mailing list since that reaches more people. You can sign up to the list on: http://www.jalview.org/mailman/listinfo/jalview-discuss
All the best,
Geoff.
On 06/07/2016 20:56, Ashok, Maitreyi (Maitreyi) wrote:
Dear Dr. Barton,
This is Maitreyi Ashok, a student at Caltech. I am working on a GPCR web server, where I want to incorporate the JalviewLite alignment viewer. I am having an issue integrating the applet in the webpage (http://abrollab.org/cgi-bin/align.py?accNum1=P07550&accNum2=O14718&Search=Submit+Query).
Can you please let me know who to direct my question towards?
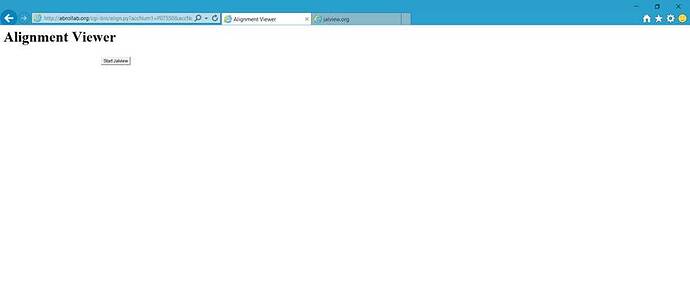
I am using Python CGI script, and have downloaded both the JalviewLite package and Java on my server. My script is below. I don’t get any explicit errors when running it, but when I open the page in Internet Explorer, the Java loading symbol comes and then a button with Start Jalview shows up. However, nothing happens when I click the button. I used the examples on http://www.jalview.org/examples/applets.html to start out with my script.
Can you please advise me on what I am doing wrong?
Thanks in advance,
Maitreyi
What the page looks like:
The script I am using:
#! /usr/bin/env python
import cgi, csv
def findSeq(prot1, num):
align1 = ‘’
globalAlign = open(‘…/gpcrs/gross-alignment.fasta’, ‘r’)
for line in globalAlign:
if ‘>’ in line and align1 != ‘’:
return align1
elif align1 != ‘’:
align1 += line
elif prot1 in line:
align1 += line
globalAlign.close()
if align1 == ‘’:
print ‘
Protein %s invalid
’ % numreturn ‘’
Set up the webpage to show results of a sequence search
print “Content-type:text/html\r\n\r\n”
print “”
print “”
print “Alignment Viewer”
print “”
print “”
print “
Alignment Viewer
”Retrieve the accession number entered and check that it’s not empty
form = cgi.FieldStorage()
prot1 = form.getvalue(“accNum1”)
prot1 = prot1.strip()
prot2 = form.getvalue(“accNum2”)
prot2 = prot2.strip()
seq1 = findSeq(prot1, 1)
seq2 = findSeq(prot2, 2)
print ‘’
print ‘’
print ‘’ %seq1
print ‘’ %seq2
print ‘’
print ‘’
print ‘’
print ‘’
print ‘<param name=“APPLICATION_URL” value=“http://www.jalveiw.org/services/launchApp” />’
print ‘Please make sure your browser supports Java’
print ‘’
Finish up the webpage
print “”
print “”
--
Geoff Barton | Professor of Bioinformatics | Head of Division of Computational Biology
School of Life Sciences | University of Dundee, Scotland, UK | [g.j.barton@dundee.ac.uk](mailto:g.j.barton@dundee.ac.uk)
Tel: +44 1382 385860 | [www.compbio.dundee.ac.uk](http://www.compbio.dundee.ac.uk) | twitter: @gjbarton
The University of Dundee is registered Scottish charity: No.SC015096