Hello all,
As we have found a workaround for dragging and dropping files has been fixed for Unix platforms its time to add the drag and drop annotation files onto an alignment. (You can also drag and drop your features files onto an alignment)
Once you have created an annotation file, just drag it onto an open alignment window in Jalview.
I've attached an image of an annotated annotation file! I'll add some proper instructions to the Wiki when I've seen peoples comments.
The test version of Jalview which reads these annotation files can be started with the following link
http://www.compbio.dundee.ac.uk/~andrew/JalviewWebsite/webstart/JalviewX.jnlp
Andrew
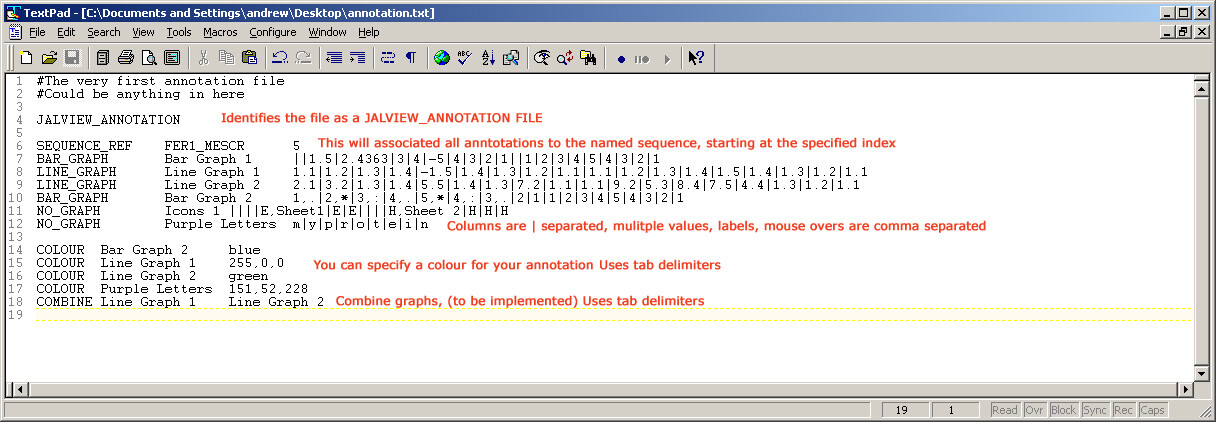
Apologies for the typos in the previously attached image, I was trying to get it done quickly before lunch.
I've added the actual text file to this email.
So to confirm the essential characteristics of the file.
The file MUST have the JALVIEW_ANNOTATION line at the top.
Each annotation row has the GRAPH_TYPE and Label at the beginning, separated with tabs. Then the column data is separated with '"|", multiple information in a column is comma separated.
The SEQUENCE_REF line is optional, otherwise the data is not associated to any particular sequence, just the whole alignment.
Andrew
annotation.txt (681 Bytes)
···
At 12:27 20/01/2006, Andrew Waterhouse wrote:
Hello all,
As we have found a workaround for dragging and dropping files has been fixed for Unix platforms its time to add the drag and drop annotation files onto an alignment. (You can also drag and drop your features files onto an alignment)
Once you have created an annotation file, just drag it onto an open alignment window in Jalview.
I've attached an image of an annotated annotation file! I'll add some proper instructions to the Wiki when I've seen peoples comments.
The test version of Jalview which reads these annotation files can be started with the following link
http://www.compbio.dundee.ac.uk/~andrew/JalviewWebsite/webstart/JalviewX.jnlp
Andrew
_______________________________________________
Jalview-dev mailing list
Jalview-dev@jalview.org
http://www.jalview.org/mailman/listinfo/jalview-dev
OK. So how do you tell it to associate graphs with different sequences? Do you just give another SEQUENCE_REF?
G.
···
On Fri, 20 Jan 2006, Andrew Waterhouse wrote:
Apologies for the typos in the previously attached image, I was trying to get it done quickly before lunch.
I've added the actual text file to this email.
So to confirm the essential characteristics of the file.
The file MUST have the JALVIEW_ANNOTATION line at the top.
Each annotation row has the GRAPH_TYPE and Label at the beginning, separated with tabs. Then the column data is separated with '"|", multiple information in a column is comma separated.
The SEQUENCE_REF line is optional, otherwise the data is not associated to any particular sequence, just the whole alignment.
Andrew
At 12:27 20/01/2006, Andrew Waterhouse wrote:
Hello all,
As we have found a workaround for dragging and dropping files has been fixed for Unix platforms its time to add the drag and drop annotation files onto an alignment. (You can also drag and drop your features files onto an alignment)
Once you have created an annotation file, just drag it onto an open alignment window in Jalview.
I've attached an image of an annotated annotation file! I'll add some proper instructions to the Wiki when I've seen peoples comments.
The test version of Jalview which reads these annotation files can be started with the following link
http://www.compbio.dundee.ac.uk/~andrew/JalviewWebsite/webstart/JalviewX.jnlp
Andrew
_______________________________________________
Jalview-dev mailing list
Jalview-dev@jalview.org
http://www.jalview.org/mailman/listinfo/jalview-dev
------------------
Geoff Barton, Professor of Bioinformatics, School of Life Sciences
University of Dundee, Scotland, UK. geoff@compbio.dundee.ac.uk
Tel:+44 1382 385860/345843 (Fax:345764) www.compbio.dundee.ac.uk
...by that I mean multiple graphs. i.e. graphA is associated with sequence 1, graphB with sequence 2 etc.
Geoff.
···
On Fri, 20 Jan 2006, Geoff Barton wrote:
OK. So how do you tell it to associate graphs with different sequences? Do you just give another SEQUENCE_REF?
G.
On Fri, 20 Jan 2006, Andrew Waterhouse wrote:
Apologies for the typos in the previously attached image, I was trying to get it done quickly before lunch.
I've added the actual text file to this email.
So to confirm the essential characteristics of the file.
The file MUST have the JALVIEW_ANNOTATION line at the top.
Each annotation row has the GRAPH_TYPE and Label at the beginning, separated with tabs. Then the column data is separated with '"|", multiple information in a column is comma separated.
The SEQUENCE_REF line is optional, otherwise the data is not associated to any particular sequence, just the whole alignment.
Andrew
At 12:27 20/01/2006, Andrew Waterhouse wrote:
Hello all,
As we have found a workaround for dragging and dropping files has been fixed for Unix platforms its time to add the drag and drop annotation files onto an alignment. (You can also drag and drop your features files onto an alignment)
Once you have created an annotation file, just drag it onto an open alignment window in Jalview.
I've attached an image of an annotated annotation file! I'll add some proper instructions to the Wiki when I've seen peoples comments.
The test version of Jalview which reads these annotation files can be started with the following link
http://www.compbio.dundee.ac.uk/~andrew/JalviewWebsite/webstart/JalviewX.jnlp
Andrew
_______________________________________________
Jalview-dev mailing list
Jalview-dev@jalview.org
http://www.jalview.org/mailman/listinfo/jalview-dev
------------------
Geoff Barton, Professor of Bioinformatics, School of Life Sciences
University of Dundee, Scotland, UK. geoff@compbio.dundee.ac.uk
Tel:+44 1382 385860/345843 (Fax:345764) www.compbio.dundee.ac.uk
_______________________________________________
Compbio mailing list
Compbio@compbio.dundee.ac.uk
http://www.compbio.dundee.ac.uk/mailman/listinfo/compbio
------------------
Geoff Barton, Professor of Bioinformatics, School of Life Sciences
University of Dundee, Scotland, UK. geoff@compbio.dundee.ac.uk
Tel:+44 1382 385860/345843 (Fax:345764) www.compbio.dundee.ac.uk
I would think that would need to be a separate annotation file to keep things simple. At present all the annotations and graphs in a file would be would be associated with the single sequence specified by SEQUENCE_REF.
···
At 13:59 20/01/2006, Geoff Barton wrote:
...by that I mean multiple graphs. i.e. graphA is associated with sequence 1, graphB with sequence 2 etc.
Geoff.
On Fri, 20 Jan 2006, Geoff Barton wrote:
OK. So how do you tell it to associate graphs with different sequences? Do you just give another SEQUENCE_REF?
G.
On Fri, 20 Jan 2006, Andrew Waterhouse wrote:
Apologies for the typos in the previously attached image, I was trying to get it done quickly before lunch.
I've added the actual text file to this email.
So to confirm the essential characteristics of the file.
The file MUST have the JALVIEW_ANNOTATION line at the top.
Each annotation row has the GRAPH_TYPE and Label at the beginning, separated with tabs. Then the column data is separated with '"|", multiple information in a column is comma separated.
The SEQUENCE_REF line is optional, otherwise the data is not associated to any particular sequence, just the whole alignment.
Andrew
At 12:27 20/01/2006, Andrew Waterhouse wrote:
Hello all,
As we have found a workaround for dragging and dropping files has been fixed for Unix platforms its time to add the drag and drop annotation files onto an alignment. (You can also drag and drop your features files onto an alignment)
Once you have created an annotation file, just drag it onto an open alignment window in Jalview.
I've attached an image of an annotated annotation file! I'll add some proper instructions to the Wiki when I've seen peoples comments.
The test version of Jalview which reads these annotation files can be started with the following link
http://www.compbio.dundee.ac.uk/~andrew/JalviewWebsite/webstart/JalviewX.jnlp
Andrew
_______________________________________________
Jalview-dev mailing list
Jalview-dev@jalview.org
http://www.jalview.org/mailman/listinfo/jalview-dev
------------------
Geoff Barton, Professor of Bioinformatics, School of Life Sciences
University of Dundee, Scotland, UK. geoff@compbio.dundee.ac.uk
Tel:+44 1382 385860/345843 (Fax:345764) www.compbio.dundee.ac.uk
_______________________________________________
Compbio mailing list
Compbio@compbio.dundee.ac.uk
http://www.compbio.dundee.ac.uk/mailman/listinfo/compbio
------------------
Geoff Barton, Professor of Bioinformatics, School of Life Sciences
University of Dundee, Scotland, UK. geoff@compbio.dundee.ac.uk
Tel:+44 1382 385860/345843 (Fax:345764) www.compbio.dundee.ac.uk
_______________________________________________
Compbio mailing list
Compbio@compbio.dundee.ac.uk
http://www.compbio.dundee.ac.uk/mailman/listinfo/compbio
Ah, I see. So for 10 sequences we would need 10 annotation files?
G.
···
On Fri, 20 Jan 2006, Andrew Waterhouse wrote:
I would think that would need to be a separate annotation file to keep things simple. At present all the annotations and graphs in a file would be would be associated with the single sequence specified by SEQUENCE_REF.
